Mechanism
You can control this model
Based on the Miura Folding, we designed a planar DNA origami (Fig.1) which can fold and transform into open and closed states.
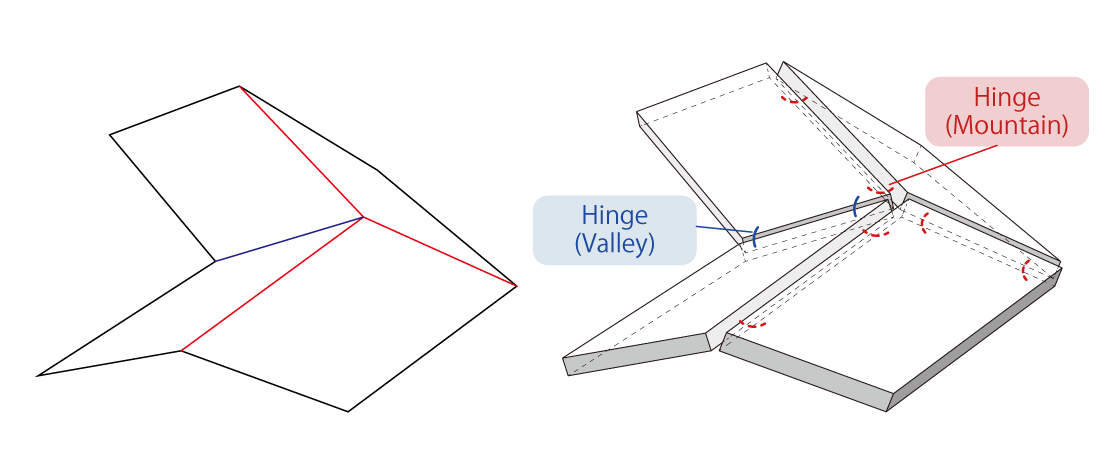
Fig.1 Miura Folding using paper (left) Miura Folding using DNA Origami (right)
Our designed structure is composed of multiple parallelogram panels made of double stranded DNA. Four panels compose a single unit, and the panels are connected by flexible single-stranded DNA called “Hinge” at the edges. As seen in the figure, Hinge are designed to be attached to the top and bottom sides of the panels so they can form mountain folds and valley folds. (Note: In order to realize smooth transformation, the four panels do not take a complete planar state so as to leave enough space to perform branch migration reactions.)
However, the main goal of our project is to transform DNA nanostructures into arbitrary forms. If we were to just simply adopt Miura Folding to DNA nanostructures, it would only form a planar structure which expands and contracts randomly. Thus, in order to control the transformation of the Miura-folded DNA Origami, we decided to confine the tiles into two states: the closed state and open state, which can be said to be much similar to the mechanism of an umbrella.
Fig.2 Umbrella Open/Closed States
As shown in the figure, for umbrellas, a mechanism is applied to fix the structure in its open and closed states. Due to this, once the umbrella takes either the open or closed state, it becomes difficult to transition to the other state. Likewise to the umbrella, we thought of a mechanism to confine the Miura-folded DNA Origami into either its open or closed state as well as enable the DNA nanostructures to transform according to specific signals. Here, we designed DNA components: “Bridge” which will hold open the DNA tiles and “Latch” which will lock the tiles shut, as well as additional signals “Close” and “Anti-close”, “Open” and “Anti-open”, which can react to regulate the transformation process (Fig.3).
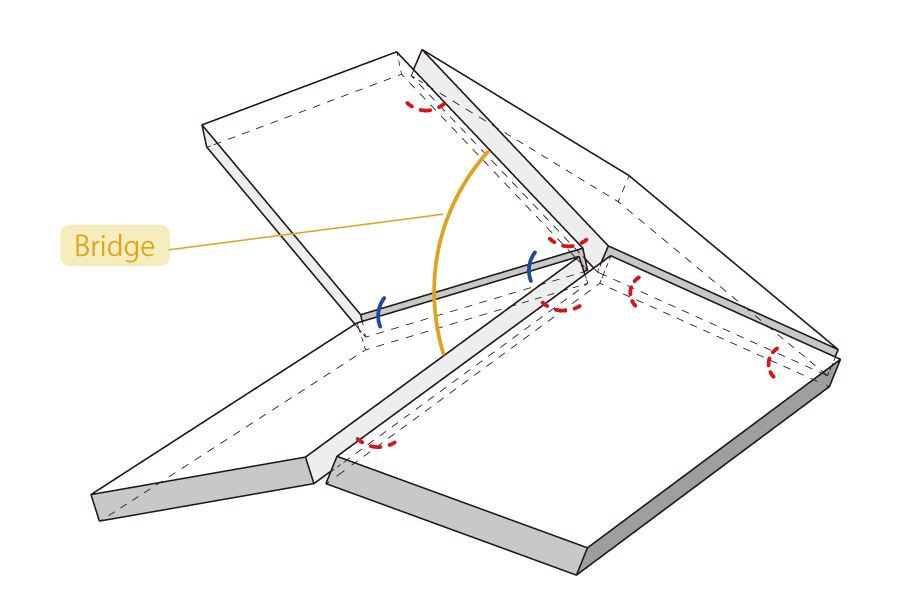
Fig3 Miura Folding using DNA Origami
As shown in (Fig.4), from the initial Neutral state, upon the addition of Close, and hybridization occurs and Latch forms double-strands with Close. This will pull the panels closer together and maintain the closed structure. To open the unit from its Closed state, first, Anti-close is added to displace Close and return state Neutral. Next, when Open which are DNA strands partially complementary to Bridge are added, hybridization occurs to form rigid double strands. This strut-like strand will hold the panels of the unit open. To once again close the unit, Anti-open is added to displace Open and return to Neutral, and further adding Close will maintain the Closed state.
Thus, the unit can open and closed repeatedly due to strand displacement reaction, and transformation can be regulated by the multiple DNA signals.
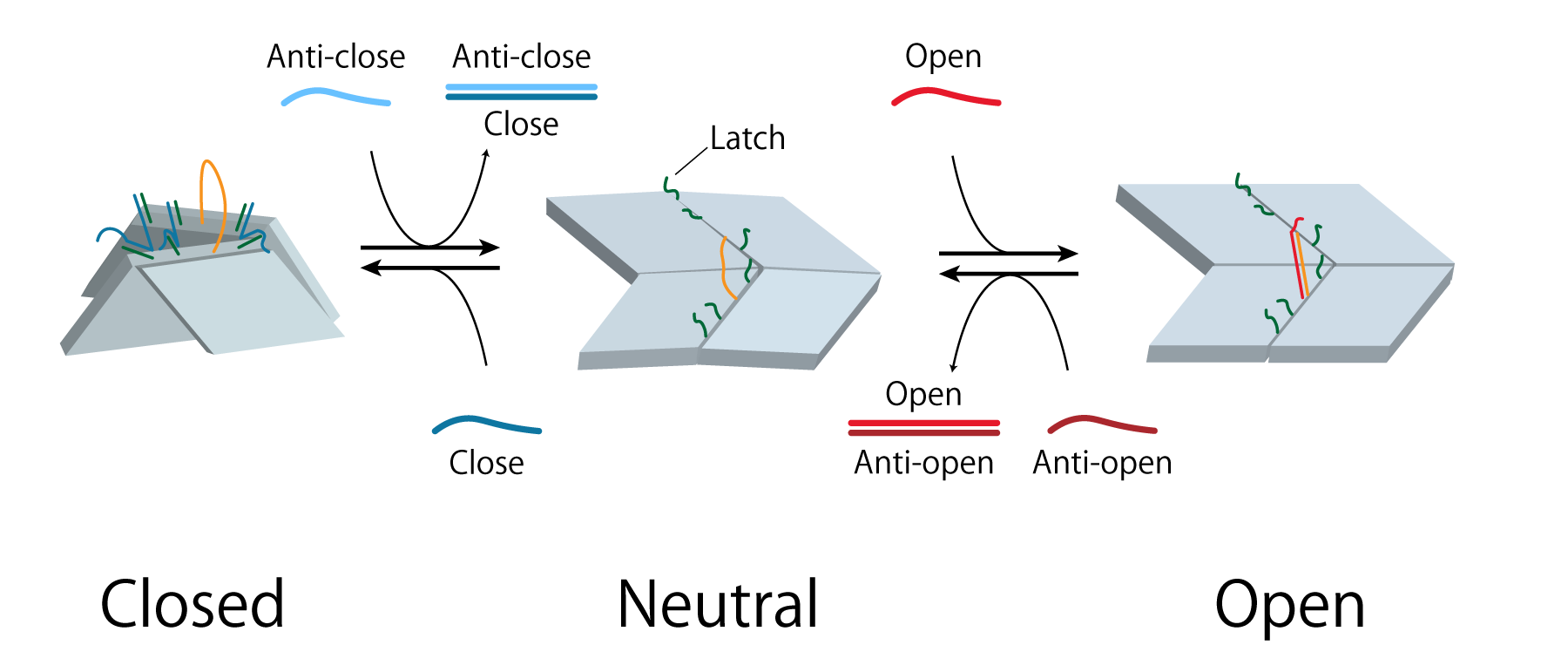
Fig.4 Mechanism of designed structure
Furthermore, by assembling multiple units, we will create a large Miura folding. Since Miura folding is a mechanism with one degree of freedom, the interlocked units will transform in unison as a whole. Therefore, by repeating our designed units, larger nanostructures can be formed and transform controllably using the above mechanism.
For those who want to know more about Miura Folding, click the following link.
caDNAno
In order to realize Miura Folding using DNA Origami, as shown in Fig.3, units made of four parallelogram panels were made from a single DNA Origami.
The structure is composed of the panels and additional DNAs called Hinge, Latch, and Bridge. Here, the scaffold and staple sequences were designed using caDNAno (Fig.5). The lattice type of the DNA Origami section was set to the square, since there was a need to accurately design the angles of the parallelograms for the DNA Origami used in our project. Compared to honeycomb, square could provide a more uniformly arranged parallelogram.
Single-stranded circular M13 bacteriophage DNA (7249 bases) were used for the scaffold, while 10-57 bases (34 bases on average) were used for the staples, and single-stranded DNA of 98 bases were used for actuator-DNA, a total of 203 DNA strands. The model created based on the caDNAno design is shown in (Fig. 6).
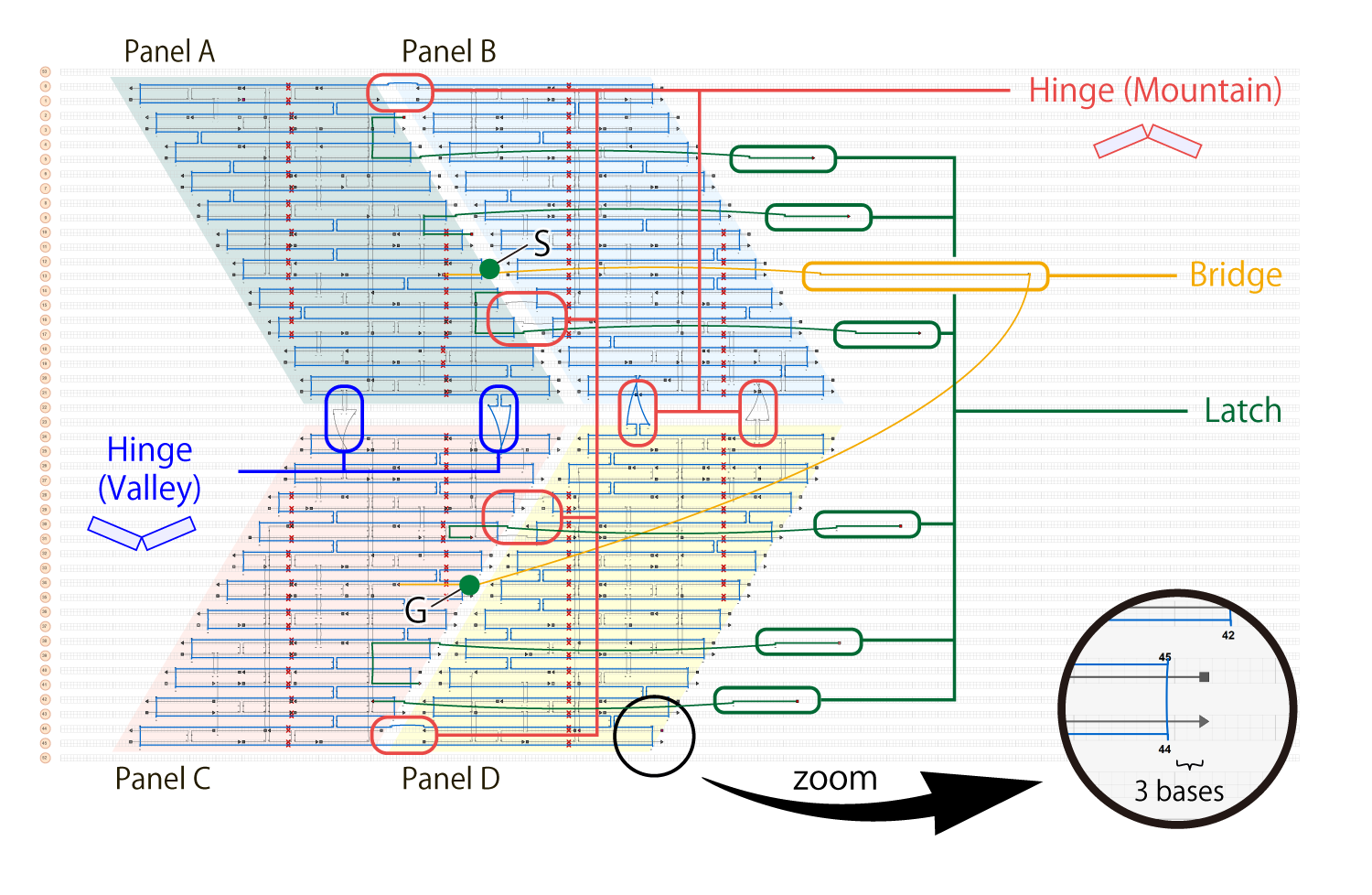
Fig.5 caDNAno design
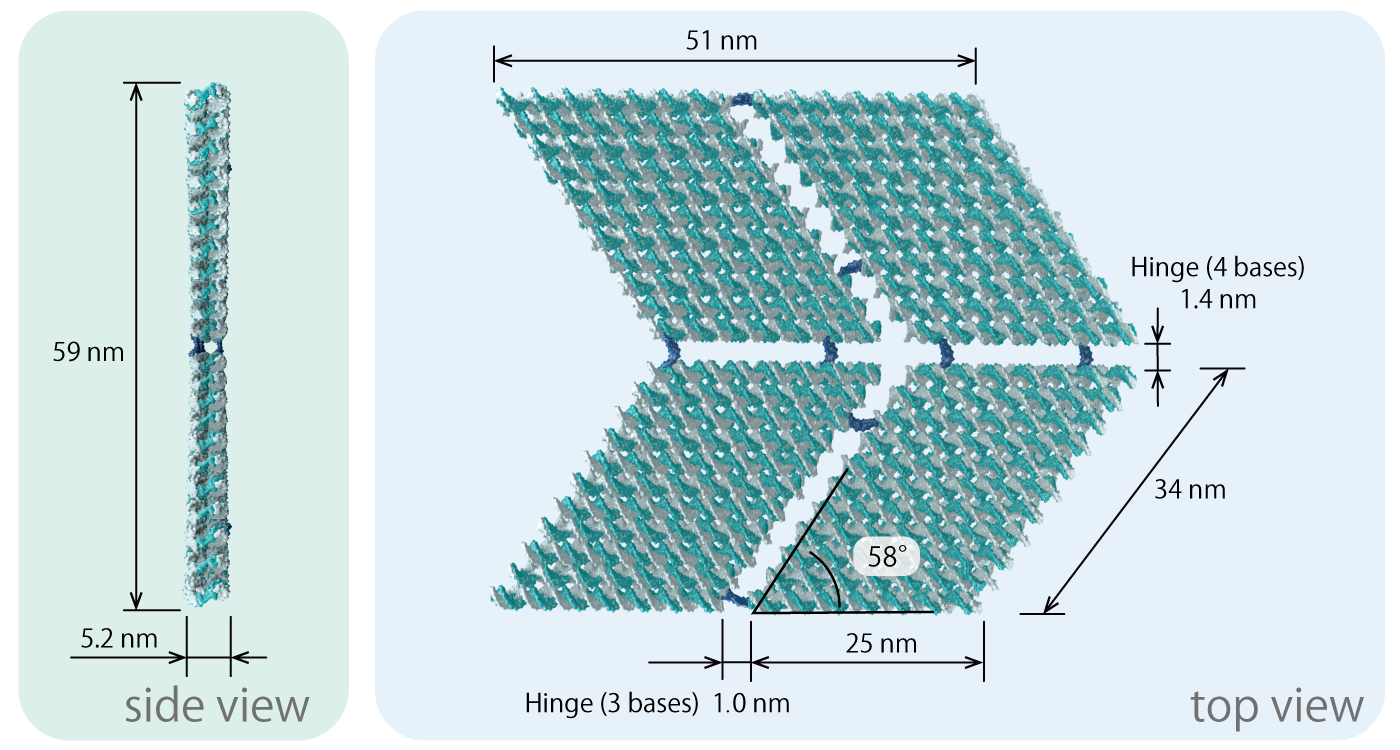
Fig.6 3D origami model
Panel
Each panel in the unit is 25 nm in width, 34 nm in length, 5.2 nm in thickness, and is made of two layers of DNA Origami. We set the angle of the parallelogram to be approximately 58 degrees. We extended the staples by three bases at the edges of the panel to avoid unwanted stacking between the edges of the panels ((Fig.5) zoomed part).
The size of the DNA Origami was calculated by setting a single DNA base to be 0.34 nm, and the average distance between neighboring DNA forming double strands as 2.6 nm.
Hinge
Panels (A, B, C, D) are connected with single-stranded DNA called Hinge. The locations of Hinge dictate the folding direction: mountain fold (shown in red in (Fig.5)) or valley fold (shown in blue in (Fig.5)).
Hinge that connect panels A-C, B-D, are 4-bases long (1.4 nm), and are attached to the upper side of the panel extending upwards, as shown in the Origami section in (Fig.7). These function as valley folding hinges. On the other hand, when DNA is extended from the lower side of the panel, the DNA functions as a mountain folding hinges.
Hinge that connect the panels A-B and C-D are 3-bases long (1.0 nm), and are attached to the upper side of the panel, extending in parallel as seen in Fig.8 (a). However, for Hinge located at the far ends of the units, in order to realize a unicursal scaffold, there is a need to extend DNA from both the upper and lower sides of the panel. Thus, as shown in Fig.8 (b), in addition to hinge-DNA extended from the bottom layer (3 bases), DNA connecting the upper layer were designed (12 bases). Here the difference in length prevents panels from interfering with the folding.
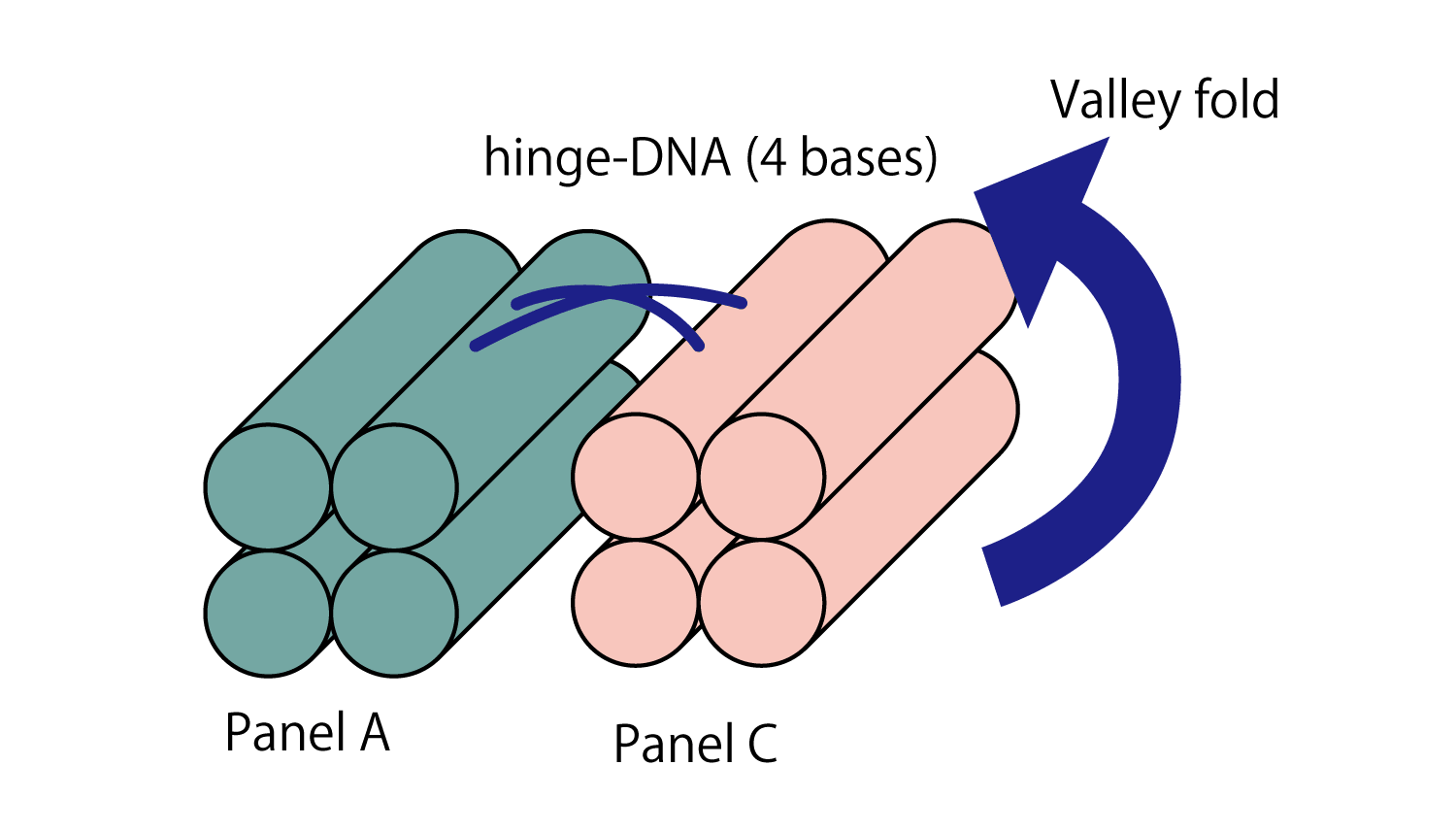
Fig.7 hinge-DNA (connecting A-C, B-D)
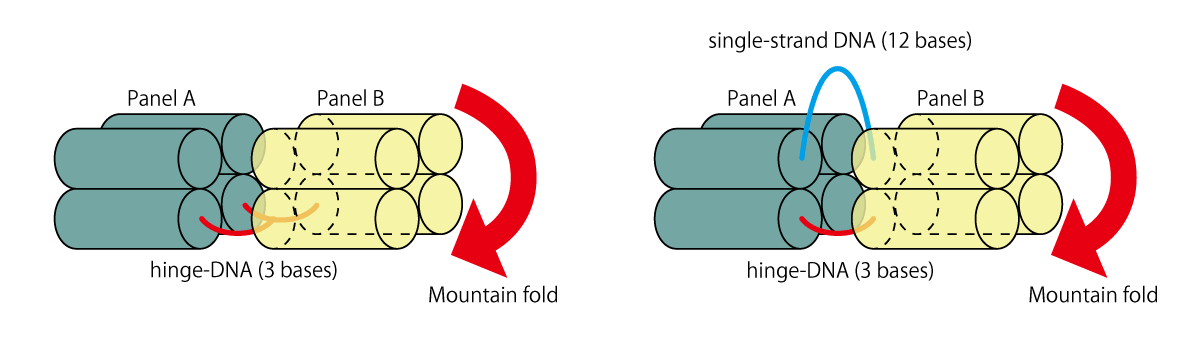
Fig.8 (a) (b) hinge-DNA (A-B, C-D)
Latch
Latch are 3 sets of single-stranded DNA attached the edges of the panels at 6 places. Upon the addition of the DNA signal, Close, Latch hybridizes with Close and its opposite ends are pulled closer to shut the panels. Its length is 21 nt. By adding this closing mechanism to the center part of the panels, formation of unwanted dimers can be reduced upon the addition of Close.
Bridge
Bridge is a single-stranded DNA extending from point S to point G in. It must be long enough to be able to open the panels when it hybridizes with Open to form a double strand. When converted into bases, the distance between point S and point G is 70 mer (23.8 nm). We designed Bridge to be slightly shorter (66 bases, 22.4 nm) since we did not want to completely open the panels.